BD Rhapsody
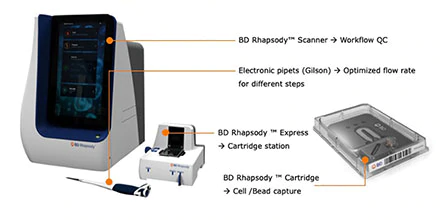
The BD Rhapsody system is able to capture thousands of individual cells in small micro-wells, enabling capture and barcoding of RNA transcripts and antibody conjugated oligos from single cells. The BD Rhapsody system offers several quality control steps during the cell capture process, revealing key parameters as cell viability, cell capture rate, number of doublets and bright field pictures of the captured cells. See Hasi Patels presentation of Rhapsody and some potential way of using Rhapsody.
We now have an updated version with 8 tests per cartridge, where you can process 8 samples together, or just use one test at the time.
Single cell RNAseq specifications
| Cells/cartridge | 100-80.000 |
|---|---|
| Number of Samples/cartridge | 1-12 |
| Run time | 2-3 hours |
| Read pairs/cell (WTA) | Rec. 50.000 |
| Cell Size | Max. 30 µm |
| Cell viability | ≥60% |
| Cell capture rate | 65%-85% |
| Illumina Seq kits | Min. 2 x 50 bp v.1.5 (NovaSeq) |
Most single cell suspensions (max. cell size 30 µm) should be compatible. It is possible to test a given cell line on a reusable cartridge available at the core facility.
Multiplet rate per lane
- 2.5% @ 10.000 cell load
- 5.2% @ 25.000 cell load
- 10.2% @ 55.000 cell load
How many cells (scRNASeq)
The number of cells per sample is highly dependent on the number of cell types present in the sample and the fractional abundances of the rarest cell types. If you have an idea about the heterogeneity of the sample, you can use the calculator developed by the SATIJA LAB to estimate a minimum number of cells/sample.
Link to calculator: https://satijalab.org/howmanycells
Sequencing depth (scRNASeq)
BD recommends a minimum sequencing depth of 50.000 Read pairs/cell (Whole Transcriptome Analysis). However, as with the number of cells, the number of read pairs/cell depends several factors like biological question asked, mRNA content of the cells, transcriptional heterogeneity etc. BD Provides a sequencing calculator that can be used to determine the appropriate Novaseq sequencing kit.
Schematic guideline of the Novaseq flowcell to use at a given cell number and sequencing depth.
Information on sequencing depth/sequencing saturation (from 10x but applicable to the BD Rhapsody):
https://kb.10xgenomics.com/hc/en-us/articles/115005062366-What-is-sequencing-saturation-
Other Applications
Look at BD Biosciences web page about Rhapsody technology (look under resources) for inspiration to use the system.
Whole transcriptome gene expression analysis.
Targeted Gene expression analysis.
Cell surface marker analysis (AbSeq).
VDJ (CDR3) sequencing.
T-cell antigen specificity analysis using Immudex dCODE dextramers.
