10X Genomics Chromium Controller
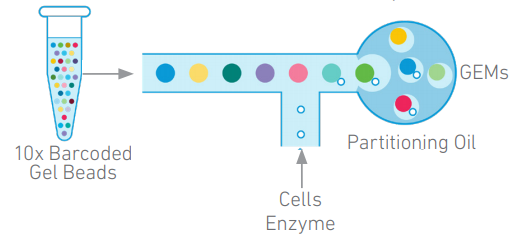
The 10X Genomics Chromium Controller is used to generate thousands of Gel bead-in-emulsion (GEMs) droplets, enabling partitioning and barcoding of single cells, HMW DNA or nuclei, in micro reactions. 10X Genomics offers several different application kits for the Chromium Controller. However, most users will probably use it for 3´ or 5´single cell RNASeq and/or single cell immune profiling. See Adam Jerauld's presentation about 10X for inspiration.
Single cell RNAseq specifications
| Cells/sample | 500-10.000 |
|---|---|
| Number of Samples/run | 1-8 |
| Run time | 18 min. |
| Read pairs/cell (3´v3.1) | Min. 20.000 |
| Cell Size | Max. 30 µm |
| Cell viability | ≥90% |
| Cell capture rate | Max. 65% |
| Illumina Seq kits | Min. 2 x 50 bp (NovaSeq) |
Most high quality single cell suspensions (max. cell size 30 µm) should be compatible. It is possible to test a given cell line on the chromium controller using a training kit.
How many cells (scRNASeq)
The number of cells per sample is highly dependent on the number of cell types present in the sample and the fractional abundances of the rarest cell types. If you have an idea about the heterogeneity of the sample, you can use the calculator developed by the SATIJA LAB to estimate a minimum number of cells/sample.
Link to calculator: https://satijalab.org/howmanycells
Sequencing depth (scRNASeq)
10X recommends a minimum sequencing depth of 20.000 Read pairs/cell (scRNASeq 3´v3.1). However, as with the number of cells, the number of read pairs/cell depends on several factors like biological question asked, mRNA content of the cells, transcriptional heterogeneity etc. More info in the links below.
Schematic guideline of the Novaseq flowcell to use at a given cell number and sequencing depth.
Information on sequencing depth/sequencing saturation:
https://kb.10xgenomics.com/hc/en-us/articles/115005062366-What-is-sequencing-saturation-
Other Applications
Single cell Copy number profiling
Reveal genome heterogeneity and clonal evolution on a single cell level.
Read more here: https://www.10xgenomics.com/products/single-cell-gene-expression
Single cell ATAC sequencing
Analyze the chromatin accessibility on a single cell level.
Read more here: https://www.10xgenomics.com/solutions/single-cell-atac/
